Maria Cuenca-Cambronero
Field and Lab Work
Field Work
I have extensive fieldwork experience in freshwater ecosystems, working in both controlled experimental settings and natural environments. I have strong experience in planning and executing large-scale field campaigns, including site selection, experimental setup, sampling design, logistics coordination, and personnel coordination. I have coordinated multidisciplinary field teams and trained students and collaborators in standardised sampling protocols, with national and international teams.
My technical field skills include the collection of physical, chemical, and biological data from freshwater habitats, focused on ponds and experimentation with micro and mesocosms. I am proficient in operating multiparameter sondes, including HOBO loggers, temperature-depth profilers, and gas flux chambers, which provide a large amount of data, which I post-process and analyse. I have experience measuring pond ecosystem functions, including greenhouse gas emissions (CO₂, CH₄, N₂O), dissolved oxygen dynamics, and ecosystem metabolism. I am also trained in the collection of aquatic divesity sediment by coring, macrophyte surveys and benthic and pelagic invertebrate sampling using nets, traps, and sediment grabs.
Lab Work
 During my six months visit at the laboratory of Biology of Marine Molluscs and Ecosystem Associated at the University of Caen I acquired experience on animal’s dissections, microtome-paraffin section, probe design, western blot, PCR, RNA extractions and in situ hybridization.
During my six months visit at the laboratory of Biology of Marine Molluscs and Ecosystem Associated at the University of Caen I acquired experience on animal’s dissections, microtome-paraffin section, probe design, western blot, PCR, RNA extractions and in situ hybridization.
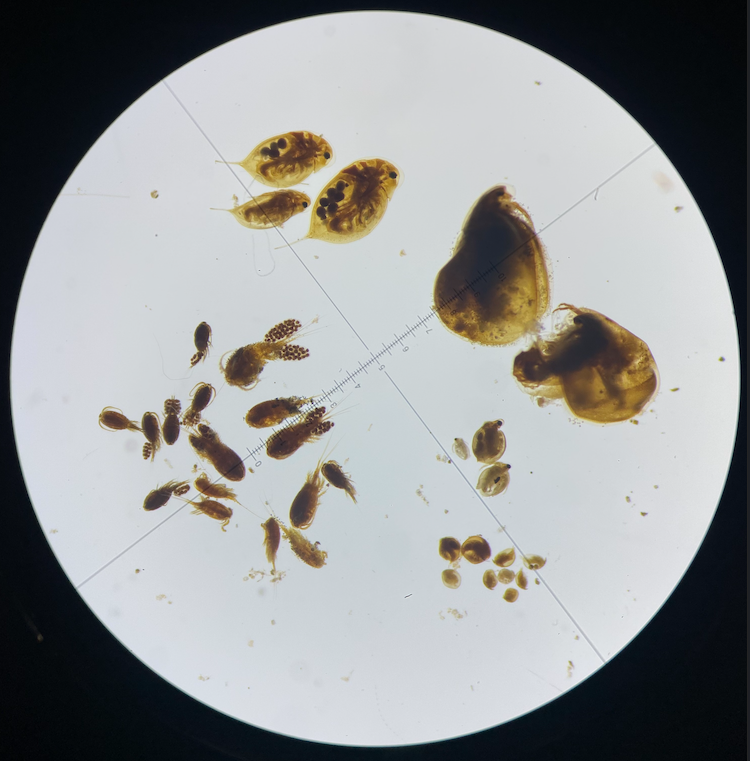
I start my PhD by learning how to work with Daphnia resting eggs (epphipia). Once I extracted the epphipia from dated sediment, I decapsulated them (Fig. 1) under the microscope, to then expose the egg to normal condition of light, temperature and nutrientes. This process increases the success of hatching eggs.
 Then, during my PhD project I oversaw protocol optimisation, DNA and RNA extraction and quantification with several molecular biology techniques (Qubit, agarose gel, qPCR, tape station and nanodrop) to optimize the sample throughput required for population genomics and functional genomics investigations including automation and miniaturization of DNA and RNA extractions, amplifications, quality assurance, and sequencing, resulting in my ability to process 500 samples simultaneously. I also performed microsatellite analysis. During my postdoc, I performed DNA extraction and protocol optimisation for de novo sequencing (Fig. 2).
Then, during my PhD project I oversaw protocol optimisation, DNA and RNA extraction and quantification with several molecular biology techniques (Qubit, agarose gel, qPCR, tape station and nanodrop) to optimize the sample throughput required for population genomics and functional genomics investigations including automation and miniaturization of DNA and RNA extractions, amplifications, quality assurance, and sequencing, resulting in my ability to process 500 samples simultaneously. I also performed microsatellite analysis. During my postdoc, I performed DNA extraction and protocol optimisation for de novo sequencing (Fig. 2).
In addition to the molecular lab skills, I have developed strong expertise in freshwater zooplankton taxonomy. I perform species-level identification of key groups, including cladocerans, copepods, and rotifers. This work requires detailed morphological knowledge and precision techniques, which often involve microscopic dissection, bleach digestion to isolate trophi (jaws) of certain rotifer species (Fig.3-5).